Course: Analyzing amplicon sequencing data with Qiime 2 - Pt. 1
Vložit
- čas přidán 20. 08. 2024
- Course: Analyzing amplicon sequencing data with Qiime 2 - Pt. 1
Christian Diener, PhD
Washington Research Foundation Distinguished Research Scientist, ISB
---------------- HARNESSING OUR INNER ECOLOGY TO TRACK AND TREAT DISEASE ----------------
A virtual course and symposium on the microbiome and its future role in precision medicine | October 15-16, 2020
ISB hosted a series of events in October 2020 that highlighted recent in silico, in vitro, and in vivo advances towards engineering the gut microbiome to resolve complex diseases. These events were virtual and free. The intended audience was graduate students, postdocs, principal investigators, industry scientists, educators, and clinicians from across the globe.
Learn about this virtual series: isbscience.org...
ISB Microbiome Slack Channel: join.slack.com...
Like ISB on Facebook: / isbsci
Follow ISB on Twitter: / isbsci








5:11 Introduction to QUIIME 2 notebook, setting up enviorment
10:45 Introduction to QUIIME alghorytm
17:07 16SrRNA amplicon sequencing vs shotgun metagenomics
21:40 Metagenomics use in diagnosing recurrent C.differens infection
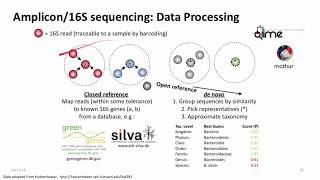
27:49 General workflow in QUIIME 16SrRNA analysis
28:44 Illumina FastQ file structure
36:03 Running first QUIMME command- importing fastq data as artifacts
41:57 Demultiplexing, creating quality visualisations of demultiplexed data
49:51 Denoising amplicon sequence variants in QUIIME
52:10 DADA2- plugin functions
54:10 DADA2- ASV Identification
57:19 DADA2- PCR Chimeras problem
1:01:42 DADA2- Merging
1:04:34 DADA2- visualisations
1:07:29 Diversity metrics- alpha and beta diversity
1:19:24 Principal Coordinate Analysis (PCoA) in phylogenetic tree construction
1:22:50 Beta diversity: 3D Emperor visualisation of PCoA
1:23:40 Statistical tests for beta diversity with PERMANOVA
1:27:57 Creating taxonomy based on sequences with Multinomial Naive Bayes
1:35:12 Final barplot visualisations
1:38:40 Creating heatmap of microbiota for samples
Thanks for sharing this! it was very helpful.
Dear Christian Diener, Thanks a lot for such an informative video presentation. It is a problem solver for me. However, I lost the track before Excercise 2. What is the code just before the Excercise 2> Secondly, can we use this program to analyze our own data?
Thanks for sharing such an informative video.
I wanted to know what things metadata.tsv file contains i.e., the column names?
important question. can we perform QIIME2 analysis in a 24 hour time for 16 samples of roughly 100K reads each.????
Question: At 49:05 you mention that cutting off the first nt´s would be conservative...what exactly do you mean by that ( gerne auch auf deutsch :-) )
And thanks so much for the upload!!!
It means the first nts have normally high quality do just remove barcode & adaper