16S vs Shotgun Metagenomic Sequencing: Pros and Cons for Microbiome Studies | Zymo Research
Vložit
- čas přidán 27. 07. 2024
- How do you choose between 16S rRNA gene sequencing and shotgun metagenomic sequencing for microbiome studies? There are many factors to consider, beyond just your budget. Learn the pros and cons of 16S and shotgun metagenomic sequencing and how your sample type and project goals can help determine which is the best option for you.
For more information on 16S vs Shotgun Metagenomic Sequencing, check out this blog post: www.zymoresearch.com/blogs/bl... - Věda a technologie



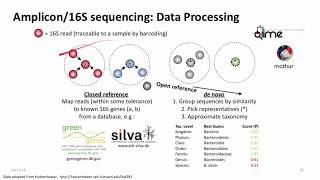





notes
- Zymo Research HostZero Microbial DNA Kit (used for shotgun sequencing)
- 16S error correction tools:
> Divisive Amplicon Denoising Algorithm 2 or DADA2, an open-source R package that improves on the DADA algorithm
> Quantitative Insights Into Microbial Ecology or QIIME (canonically pronounced 'Chime')
- 16s bioinformatic tool: PICRUSt (pronounced “pie crust”) or Phylogenetic Investigation of Communities by Reconstruction of Unobserved States, written in Python and R
Thanks for this wonderful video. 🌞
Great video! A suggestion for something you could do in the future might be to make a video comparing Amplicon Sequencing and Shotgun Sequencing, as 16S is just an example of an amplicon. You state that the use of 16S region limits your study to just bacteria, but other highly conserved regions can be used to target members of the other domains, like the 18S and ITS regions like you mentioned at the end.
Good point
Thanks for uploading ~ Great video, it provides an informative insight into the two methods.
This is a really nice video. Very helpful!
thank you this is amazing
Curious how cheap I could drive sequencing for some very low complexity population samples. I'm interested in getting full genomes for some samples that probably have 10 or less species/strains as cheap as possible.
Can you also explain to me a bit what do you mean by cross-domain coverage?
Hey Zymo Research... pls do you have in store some references discussing about metagenomics vs metabarcoding to share with us (me especially)? Thanks by advance!
Why did you compare to QIIME version 1.9 for the false positive rate? Isn't there a more recent version, particularly in QIIME2?
Hi Brian! The experiment was performed before QIIME 2 was released but, the essence of this experiment is comparing OTU (QIIME) and ASV (DADA2). An upcoming video will dive into the differences between the OTU and ASV approaches.
Why are there no error correction tools for shotgun sequencing?
My guess is that the tests go through a correction algorithm, and are already factored in.